Modern methodologies of molecular biology allow transference of foreign genes into major crop plants. In BarMatinTM the thaumatin gene was transferred with the gene shuttle Agrobacterium tumefaciens. The principle of that approach is illustrated and described in figure 1 (see Gelvin, (2003) Improving plant genetic engineering by manipulating the host. Trends Biotechnol (21): 95-98). For the transformation, the barley line Golden Promise was used because it was known to be efficiently infectable with Agrobacterium tumefaciens (see Orman-Ligeza et al., (2020) TRA1: A locus responsible for controlling Agrobacterium-mediated transformability in barley. Front Plant Sci (11): 1-11).

Figure 1: Principle of the integration of the thaumatin gene into the genome of barley (red insert). The gene was originally cloned from katamfe and was subsequently inserted together with an issue-specific promoter into a Ti-plasmid. This artificial plasmid construct is transformed into Agrobacterium tumefaciens by electroporation. After the artificial bacterium has infected the host cell, the gene is transferred in a randomized chromosomal position into the barley genome. The transformation process is a complex interaction of the virulent proteins of the gene shuttle with the barley host cell. Due to the co-transformation of the bar herbicide resistance gene, a stringent selection of cells with integrated thaumatin could be conducted. The thaumatin gene in BarMatinTM is homozygously present and, according to that, the line could be stably preserved and simply propagated.
The native thaumatin protein has a length of 207 amino acids and molecular mass of 22 kDa. The isoforms I and II have indeed the same length; however, they differ on the positions of four amino acids. The three-dimensional (3D) structure of thaumatin I consist of three domains (i–iii), in which domains ii and iii are characterized by several disulphide bridges, ensuring the protein a high temperature stability. In total, the protein has eight of such covalent bridges and the 3D structure of the protein is the company logo of BarMatin UG (see De Vos et al., (1985) Three-dimensional structure of thaumatin I, an intensely sweet protein. Proc Natl Acad Sci USA (82): 1406-1409).
BarMatinTM uses a known storage protein promoter sequence, which is located 5‘-upstream of the gene start codon. This combination ensures that the thaumatin protein is synthesized in large amounts on the ribosomes of the endoplasmic reticulum of cells of endosperm of the kernel, as it is normally done analogously with the storage proteins that the kernel produces for seedling nutrition in nature. Specific transcription factors present solely in the endosperm leads to switching on the gene only during the kernel ripening period. The thaumatin gene, therefore, is not active in other plant parts such as leaves, stem or roots (see figure 2).
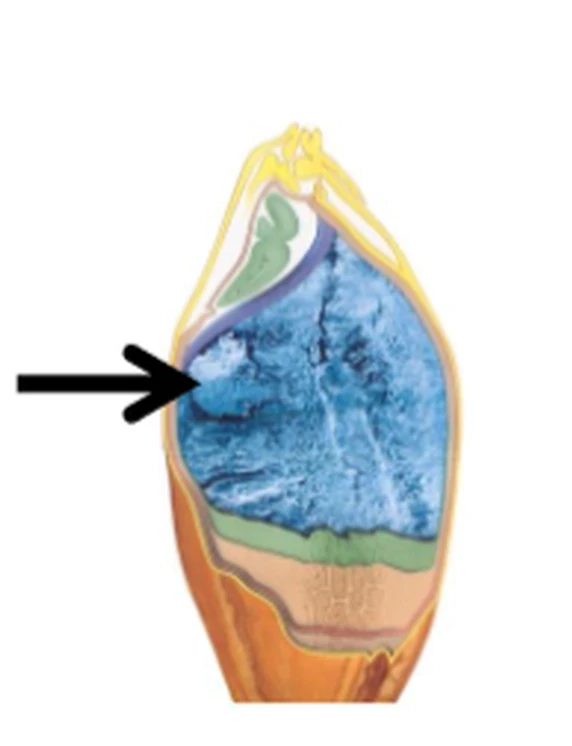
Figure 2: Longitudinal view of a barley kernel with blue coloured endosperm. In BarMatinTM the accumulation of thaumatin takes place specifically there (arrow indicating the position of synthesis).
In the past, the presence of thaumatin protein in BarMatinTM was verified by using different methodologies. For example, the genomic integration and the correct transcription of mRNA could be confirmed by PCR-based techniques. At the level of protein translation, it could be demonstrated that BarMatinTM was able to produce the identical thaumatin protein I as that was available from katamfe (protein sequencing by Edman degradation). Electrophoretic separation of purified thaumatin-protein BarMatinTM brings about a distinct 22 kDa band (see figure 3).

Figure 3: Differently purified thaumatin I extracted from kernels of BarMatinTM and electrophoretically separated by the use of a polyacrylamide gel and stained with Coomassie blue. The purified protein isolated from kernels of BarMatinTM (Lanes 4, 3, 1 and 0.5) belongs to a 22 kDa band that exactly corresponds to the molecular mass of thaumatin I (see black arrow). One kilogramme of kernels can produce 3 g of purified thaumatin on average. It may be mentioned here that protein content in the kernels strongly depends on the nitrate supply of the plants.
Even a more explicit proof of the identity of thaumatin I protein in BarMatinTM provides the so called peptide maps of enzymatic degradation products that can be separated chromatographically. The resulting patterns are entirely identical for BarMatinTM and katamfe (see figure 4).
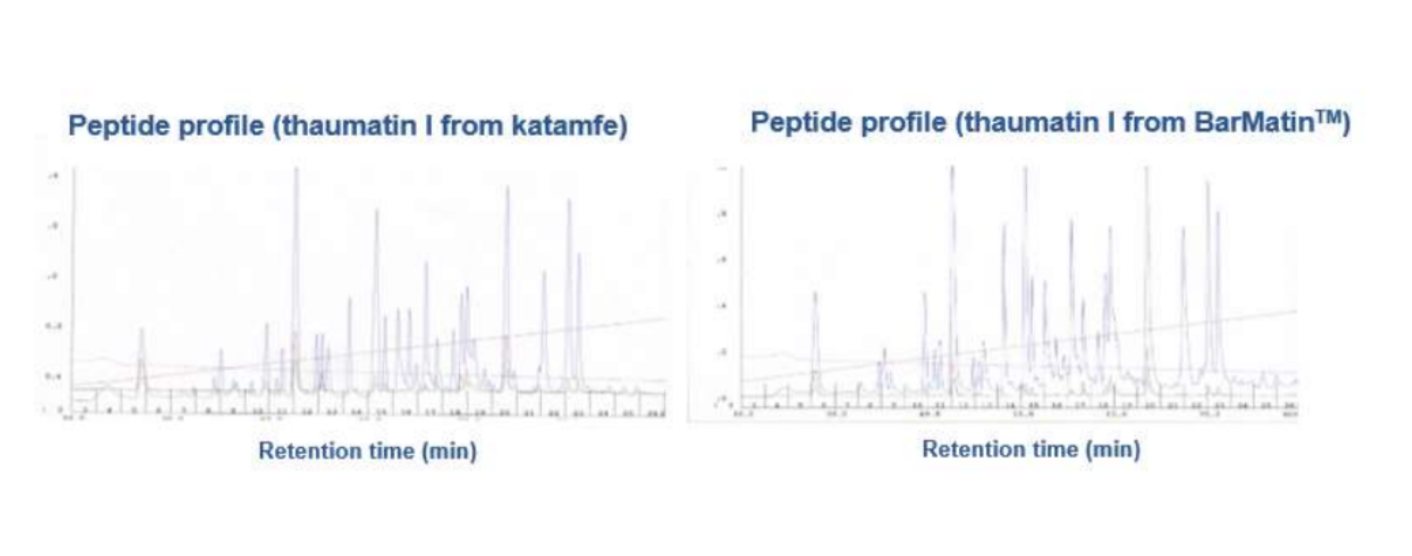
Figure 4: Enzymatic degradation of thaumatin I from katamfe (left chromatogram) and BarMatinTM (right chromatogram).
Even simple eating of a kernel provides the proof: BarMatinTM accumulates thaumatin I in huge quantities!
You are coming from the food, plant breeding, beverage or pharmaceutical industry and have interest on BarMatinTM?
Contact us soon!
We answer to all inquiries immediately and discretely! We are pleased for your interest!